News about the lab
NERVSPAN - Molecular plasticity of neurons during C. elegans lifespan
2024-05 - Marie Skłodowska-Curie Actions - European Training Network


Within the framework of the NERVSPAN European Doctoral Training Network, we are offering a fully-funded PhD fellowship to study the robustness of neuronal excitability and function across gender and lifespan.
The intrinsic electrical properties of neurons are defined by the repertoire of ion channels they express. Determining the precise genetic programs that establish the electrical identity of neurons remains a major question in cellular neuroscience. Single-cell transcriptomic studies in the C. elegans nervous system have revealed that potassium channels are often expressed in complex combinations of over a dozen channel genes in a single neuron. These different ion channel combinations will determine the biophysical and electrical properties defining neuron-specific functions throughout its life.
From a conceptual point of view, this project will address for the first time how ion channel degeneracy, variability and covariation are genetically regulated to ensure the resilience and phenotypic stability/plasticity of excitable cells in C. elegans.
NERVSPAN will train 12 doctoral Students at 7 academic institutions and 1 company. The nature of the programme will provide the enrolled students with strong interdisciplinary training (including, but not limited to: molecular biology, bioinformatics, high resolution microscopy, proteomics, and transcriptome profiling).
New publication - A tonically active master neuron modulates mutually exclusive motor states at two timescales
2024-04-10
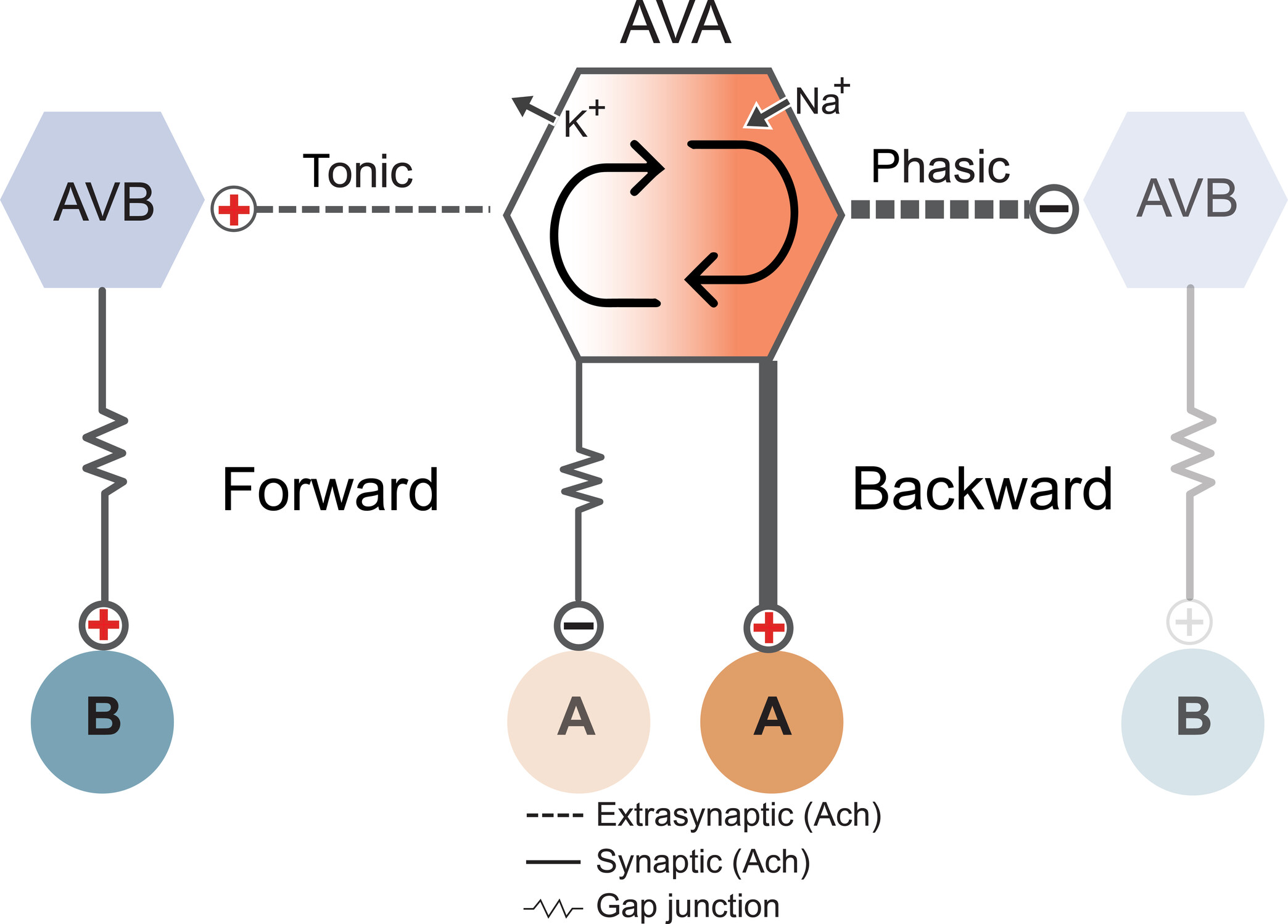 Congratulations to Jun, Sonia El Mouridi, and the teams of Mei Zhen and Shangbang Gao for our first collaborative study about the exquisite regulation of C. elegans locomotion.
Congratulations to Jun, Sonia El Mouridi, and the teams of Mei Zhen and Shangbang Gao for our first collaborative study about the exquisite regulation of C. elegans locomotion.
In this study, we report a new conceptual framework for a universal problem in motor control: how do animals generate smooth transitions when switching between opposing motor states like forward and backward locomotion? We analyzed circuits for forward and backward locomotion in C. elegans with single cell endogenous membrane potential perturbations, electrophysiology, calcium imaging, and mathematical modeling. In contrast to the prevailing view, our results establish a new model for smooth and continuous behavioral transitions between forward and backward locomotion.
New publication - Natural variation in the Caenorhabditis elegans egg-laying circuit modulates an intergenerational fitness trade-off
2024-04-02
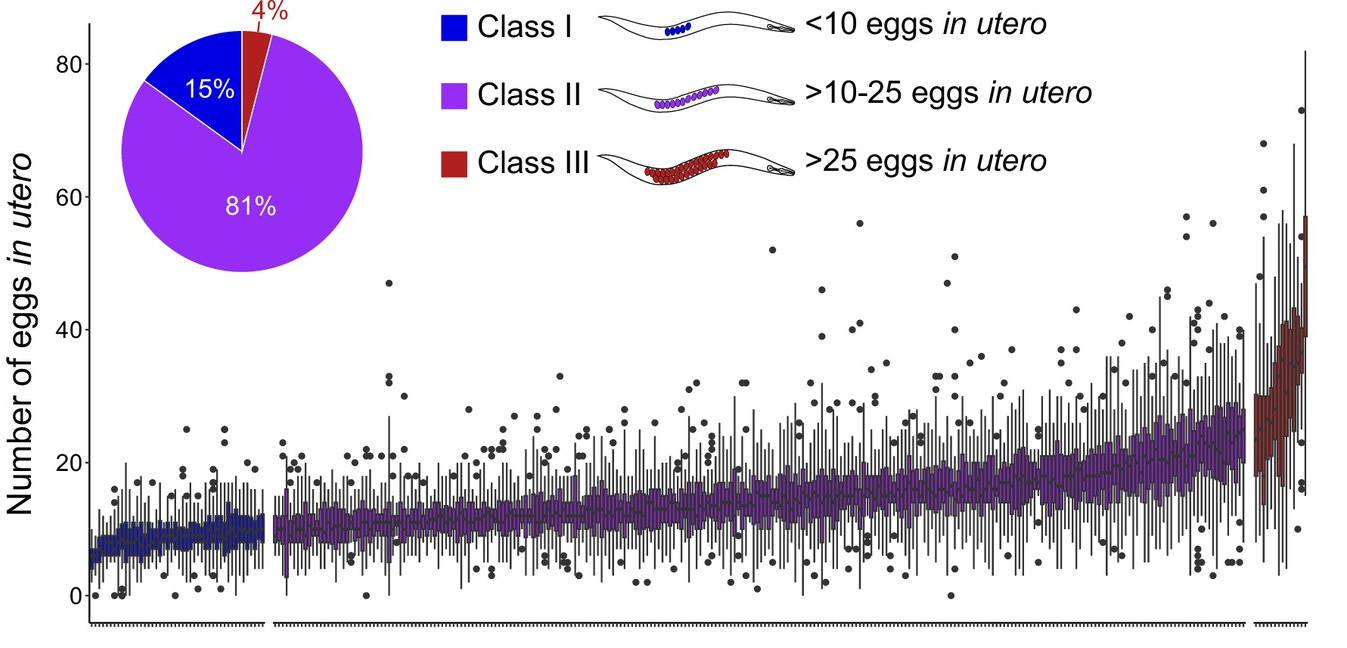 Congratulations to Laure Mignerot and the team of Christian Braendle for our second collaborative study about the natural variation of the C. elegans egg-laying behaviour.
Congratulations to Laure Mignerot and the team of Christian Braendle for our second collaborative study about the natural variation of the C. elegans egg-laying behaviour.
eLife assessment - This important work provides a thorough and detailed analysis of natural variation in C. elegans egg-laying behavior. The authors present convincing evidence to support their hypothesis that variations in egg-laying behavior are influenced by trade-offs between maternal and offspring fitness. This study establishes a framework for elucidating the molecular mechanisms underlying this paradigm of behavioral evolution.
New publication - Functional and clinical characterization of a novel homozygous KCNH2 missense variant in the pore region of Kv11.1 leading to a viable but severe long-QT syndrome
2023-12-21
 Congratulations to Antoine, Olga and the team of Philippe Chevalier (Rhythmology department, HCL Lyon) for our first collaborative study about Kv11.1/hERG potassium channels.
Congratulations to Antoine, Olga and the team of Philippe Chevalier (Rhythmology department, HCL Lyon) for our first collaborative study about Kv11.1/hERG potassium channels.
Highlights
- Homozygous missense variants in the pore of Kv11.1 often lead to intrauterine death.
- We describe a novel hERG p.Gly603Ser homozygous loss-of-function variant.
- It causes a severe but viable long-QT syndrome with a delayed clinical expression.
- Family segregation and functional analysis classify hERG p.Gly603Ser as probably pathogenic.
DYSCO - Dystrophin-associated protein complex and subcellular compartmentalization
2023-07-13

 Our project with the team of Vincent Mirouse (iGred, Clermont-Ferrand) and Helge Amthor (UVSQ) has been selected by ANR ! The Dystrophin Associated Protein Complex (DAPC) is a key actor of the cell – extracellular matrix (ECM) interface, as revealed by its implication in human genetic disorders. However, its molecular and cellular functions are still poorly understood because tractable model systems allowing state-of-the-art in vivo cell biology and genetic approaches are lacking. Our three teams have developed the first transgenic dystrophin reporters that have revealed remarkably compartmentalized membrane distributions of the DAPC in epithelia and muscle cells in C. elegans, Drosophila, and mouse. The project aims to elucidate the organization and dynamics of the DAPC and to characterize its new functions in relation to specific cell cortical compartments.
Our project with the team of Vincent Mirouse (iGred, Clermont-Ferrand) and Helge Amthor (UVSQ) has been selected by ANR ! The Dystrophin Associated Protein Complex (DAPC) is a key actor of the cell – extracellular matrix (ECM) interface, as revealed by its implication in human genetic disorders. However, its molecular and cellular functions are still poorly understood because tractable model systems allowing state-of-the-art in vivo cell biology and genetic approaches are lacking. Our three teams have developed the first transgenic dystrophin reporters that have revealed remarkably compartmentalized membrane distributions of the DAPC in epithelia and muscle cells in C. elegans, Drosophila, and mouse. The project aims to elucidate the organization and dynamics of the DAPC and to characterize its new functions in relation to specific cell cortical compartments.
Paper Alert - Distinct dystrophin and Wnt/Ror-dependent pathways establish planar-polarized membrane compartments in C. elegans muscles
2023-03-28

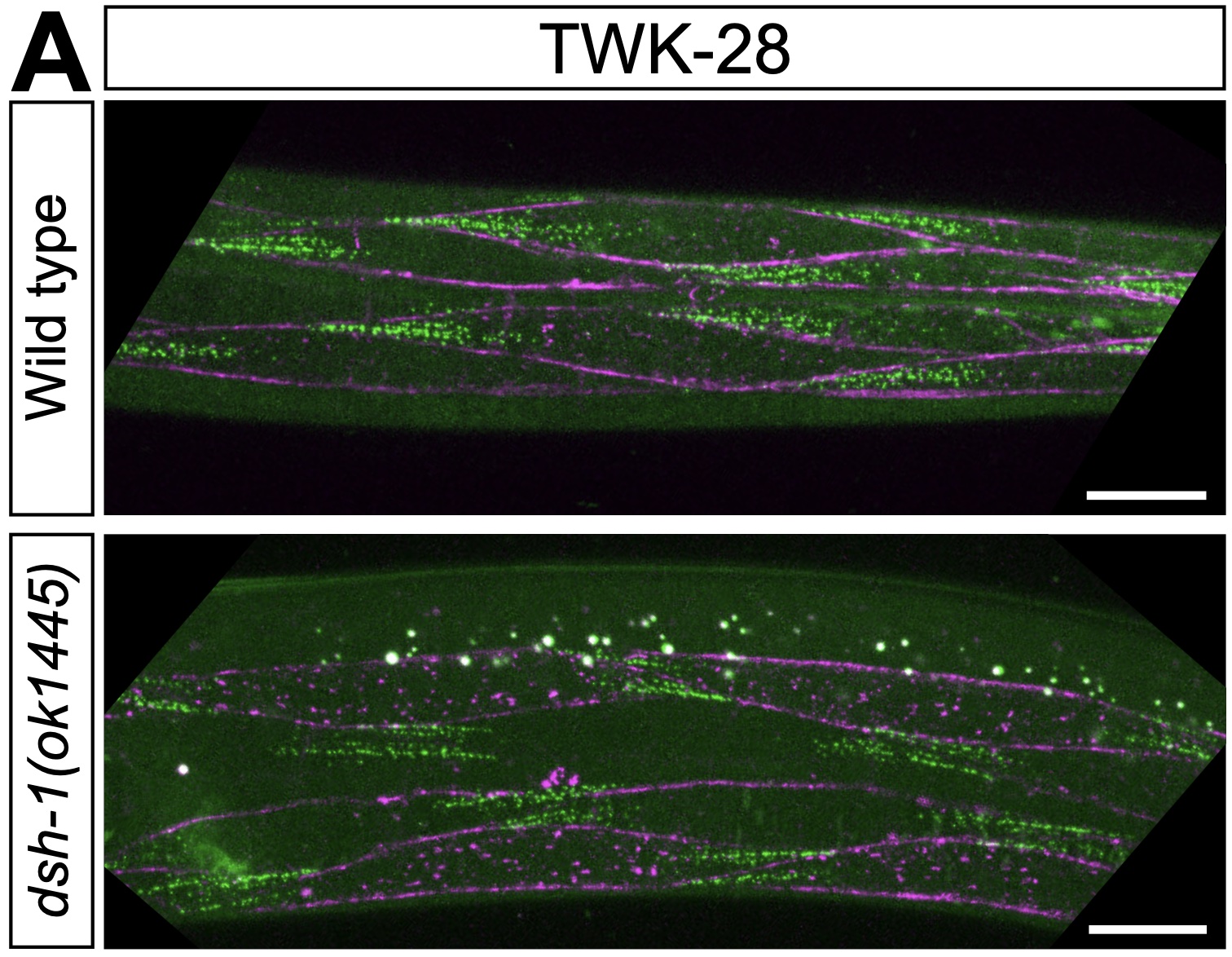 Our study revealing the remarkably complex organisation of the worm's sarcolemma is now on BioRxiv!
Our study revealing the remarkably complex organisation of the worm's sarcolemma is now on BioRxiv! Four and half years in the making, congratulations to Alice, Nora, Marie, Amandine, Noémie and Olga for this magnum opus revealing this entirely unsuspected new case of planar cell polarity in C. elegans muscles.
SUMMARY
The plasma membrane of excitable cells is highly structured and molecular scaffolds recruit proteins to specific membrane compartments. Here, we show that potassium channels and proteins belonging to the dystrophin-associated protein complex define multiple types of planar-polarized membrane compartments at the surface of C. elegans muscle cells. Surprisingly, conserved planar cell polarity proteins are not required for this process. However, we implicate a Wnt signaling module involving the Wnt ligand EGL-20, the Wnt receptor CAM-1, and the intracellular effector DSH-1/disheveled in the formation of this cell polarity pattern. Moreover, using time-resolved and tissue-specific protein degradation, we demonstrate that muscle cell polarity is a dynamic state, requiring continued presence of DSH-1 throughout post-embryonic life. Our results reveal the intricate, highly reproducible, and entirely unsuspected complexity of the worm's sarcolemma. This novel case of planar cell polarity in a tractable genetic model organism may provide valuable insight into the molecular and cellular mechanisms that regulate cellular organization, allowing specific functions to be compartmentalized within distinct plasma membrane domains.Fleggsibility - Dissecting the microevolutionary flexibility of a neural circuit
2022-08-13
 Our project with the team of Christian Braendle (iBV Nice) and ViewPoint: Behavior Analysis Technologies has been selected by ANR ! On the heals of our joint publication (Vigne et al., Science Advances 2021) we will be trying to understand how specific neural circuits evolve to generate natural behavioural variation within species. In particular, the precise genetic changes that modulate cellular and developmental architectures of reproductive systems remain unclear. Here we will focus on the simple egg-laying circuit of the nematode Caenorhabditis elegans as a powerful model system to study natural microevolutionary (intraspecific) variability.
Our project with the team of Christian Braendle (iBV Nice) and ViewPoint: Behavior Analysis Technologies has been selected by ANR ! On the heals of our joint publication (Vigne et al., Science Advances 2021) we will be trying to understand how specific neural circuits evolve to generate natural behavioural variation within species. In particular, the precise genetic changes that modulate cellular and developmental architectures of reproductive systems remain unclear. Here we will focus on the simple egg-laying circuit of the nematode Caenorhabditis elegans as a powerful model system to study natural microevolutionary (intraspecific) variability.
Now published in Molecular Genetics and Metabolism!
2021-08-09
Great collaboration with the Undiagnosed Diseases Network's Tim Schedl and Stephen Pak.
Papers never tell the “story” behind the science. So much had to happen for this to come together! From eagle-eyed curiosity-driven observations, great timing, to outstanding scientific collaboration. I feel lucky for the opportunity to bring such a meaningful work to fruition.
Kudos to all involved in Europe and the US. Past and present lab members who all contributed their essential piece.

Neurobeachin (NBEA) was initially identified as a candidate gene for autism. Recently, variants in NBEA have been associated with neurodevelopmental delay and childhood epilepsy. Here, we report on a novel NBEA missense variant (c.5899G > A, p.Gly1967Arg) in the Domain of Unknown Function 1088 (DUF1088) identified in a child enrolled in the Undiagnosed Diseases Network (UDN), who presented with neurodevelopmental delay and seizures. Modeling of this variant in the Caenorhabditis elegans NBEA ortholog, sel-2, indicated that the variant was damaging to in vivo function as evidenced by altered cell fate determination and trafficking of potassium channels in neurons. The variant effect was indistinguishable from that of the reference null mutation suggesting that the variant is a strong hypomorph or a complete loss-of-function. Our experimental data provide strong support for the molecular diagnosis and pathogenicity of the NBEA p.Gly1967Arg variant and the importance of the DUF1088 for NBEA function.
Keywords: C. elegans; Epilepsy; Neurobeachin; Neurodevelopmental delay; SEL-2; NEDEGE OMIM 619157; Two-pore domain potassium channels.
New team photo
June 2021
Welcome Uma and Marine.

Molecular, cellular, and clinical investigation of the autism, epilepsy, and neurodevelopmental disorder gene Neurobeachin/NBEA
2020-09-29


Neurobeachin is a brain-specific protein required for vesicular trafficking, synaptic structure, and synaptic targeting of neurotransmitter receptors. Neurobeachin mutations have very recently been identified as a genetic cause of autism, neurodevelopmental delay and early generalized epilepsy (Mulhern et al., 2018). During a genetic screen in C. elegans, we have discovered that Neurobeachin can also regulate potassium channel trafficking. Interestingly, these channels play a central role in controlling neuronal excitability. In this project, we propose to combine clinical and fundamental research approaches to better understand the pathophysiology of this new neurological disease, identify cellular processes regulated by Neurobeachin, and find new molecular partners that could serve as potential therapeutic targets in the future.
This project will be a collaborative venture between our team, Tristan T. Sands and Natalie Lippa (Columbia University, New York), Sarah Weckhuysen (VIB & University of Antwerp), Qiang Liu (Bargmann Lab, Rockefeller University, New York), and Maëlle Jospin (Bessereau Lab, Institut NeuroMyoGène, Lyon).
If you are interested in joining the project as a Post-doctoral fellow, Masters, or PhD Student, don't hesitate to contact us!
Now published in Science Advances!
2021-02-05
Great collaboration with Christian Braendle : from wild worms to a single point mutation in an SK potassium channel.

Genetic assimilation—the evolutionary process by which an environmentally induced phenotype is made constitutive—represents a fundamental concept in evolutionary biology. Thought to reflect adaptive phenotypic plasticity, matricidal hatching in nematodes is triggered by maternal nutrient deprivation to allow for protection or resource provisioning of offspring. Here, we report natural Caenorhabditis elegans populations harboring genetic variants expressing a derived state of near-constitutive matricidal hatching. These variants exhibit a single amino acid change (V530L) in KCNL-1, a small-conductance calcium-activated potassium channel subunit. This gain-of-function mutation causes matricidal hatching by strongly reducing the sensitivity to environmental stimuli triggering egg-laying. We show that reestablishing the canonical KCNL-1 protein in matricidal isolates is sufficient to restore canonical egg-laying. While highly deleterious in constant food environments, KCNL-1 V530L is maintained under fluctuating resource availability. A single point mutation can therefore underlie the genetic assimilation—by either genetic drift or selection—of an ancestrally plastic trait.
Newest Preprint out on BioRXiv!
2020-06-29
 “A single nucleotide change underlies the genetic assimilation of a plastic trait”
“A single nucleotide change underlies the genetic assimilation of a plastic trait”Great collaboration with Christian Braendle : from wild worms to a single point mutation in an SK potassium channel. Great how science pulls us all together!
Abstract
Genetic assimilation - the evolutionary process by which an ancestral environmentally sensitive phenotype is made constitutive - is a fundamental concept in biology. Its evolutionary relevance is debated, and our understanding of its prevalence, and underlying genetics and molecular mechanisms, is poor. Matricidal hatching is an extreme form of maternal provisioning induced by adverse conditions, which varies among Caenorhabditis elegans populations. We identified wild isolates, sampled from natural populations across multiple years and locations, that express a derived state of near-constitutive matricidal hatching. A single amino acid change in kcnl-1, encoding a small-conductance calcium-activated potassium channel subunit, explains most of this variation. A gain-of-function mutation altering the S6 transmembrane domain causes inappropriate activation of the K+ channel, leading to reduced vulval muscle excitability, and thus reduced expulsion of embryos, irrespective of environment. Using reciprocal allelic replacements, we show that this amino acid change is sufficient to induce constitutive matricidal hatching whilst re-establishing the ancestral protein abolishes matricidal hatching and restores egg-laying, thereby doubling lifetime reproductive fitness under benign conditions. While highly deleterious in the laboratory, experimental evolution showed that KNCL-1(V530L) is maintained under fluctuating resource availability. Selection on a single point mutation can therefore underlie the genetic assimilation of an ancestrally plastic trait with drastic life-history consequences.
New team photo
February 2020
Big thanks to our pre-COVID-19 interns Ingrid and Fiona. Lots of energy and exciting results.

Modeling disease-causing mutations of Neurobeachin/NBEA in the nematode Caenorhabditis elegans
2019-07-27

We are grateful to the Fondation Maladies Rares that will support our study of variants of unknown significance of Neurobeachin / NBEA. This funding will allow us to create new tools in the model nematode C. elegans to evaluate the impact of mutations causing this very rare disease associated with strong neurodevelopmental deficits and epilepsy (Mulhern et al., 2018).
We hope that this work will contribute to the diagnosis of patients with novel variants in Neurobeachin in the future in order to better define the phenotypic spectrum of this "disease" (NEDEGE).
New team photo
July 2019
A big thank you to Léa, Charlotte, Emilie (not pictured), and Quentin who have been wonderful rotation students this year. They have each pioneered new projects in which we model genetic variant for human diseases in C. elegans.

Functional evaluation of Neurobeachin/NBEA genetic variants in the model nematode Caenorhabditis elegans

2019-02-28
Today on #RareDiseaseDay we submit our 1st application to the NIH's Undiagnosed Diseases Network "Gene Function Studies" call. It flows directly from ERC-funded blue-sky research. We hope that it will get funded so we can demonstrate (once more...) how basic non-applied "worm research" leads directly to Rare Disease model systems!
2019-05-30
Despite very encouraging and positive comments, our proposal was not funded by UDN. We'll keep working on it and try again next year.
Newest paper out today!
2019-02-15
“Mutation of a single residue promotes gating of vertebrate and invertebrate two-pore domain potassium channels”
Great collaboration between our group, IPMC Nice's Delphine Bichet and Florian Lesage and Dawon Kang at Gyeongsang National University.
Editor's Summary: "Mutations that modulate the activity of ion channels are essential tools to understand the biophysical determinants that control their gating. Here authors reveal the role played by a single residue in the second transmembrane domain of vertebrate and invertebrate two-pore domain potassium channels."
Indeed, we found that “Mutation of a single residue promotes gating of vertebrate and invertebrate two-pore domain potassium channels”. We named this position TM2.6.
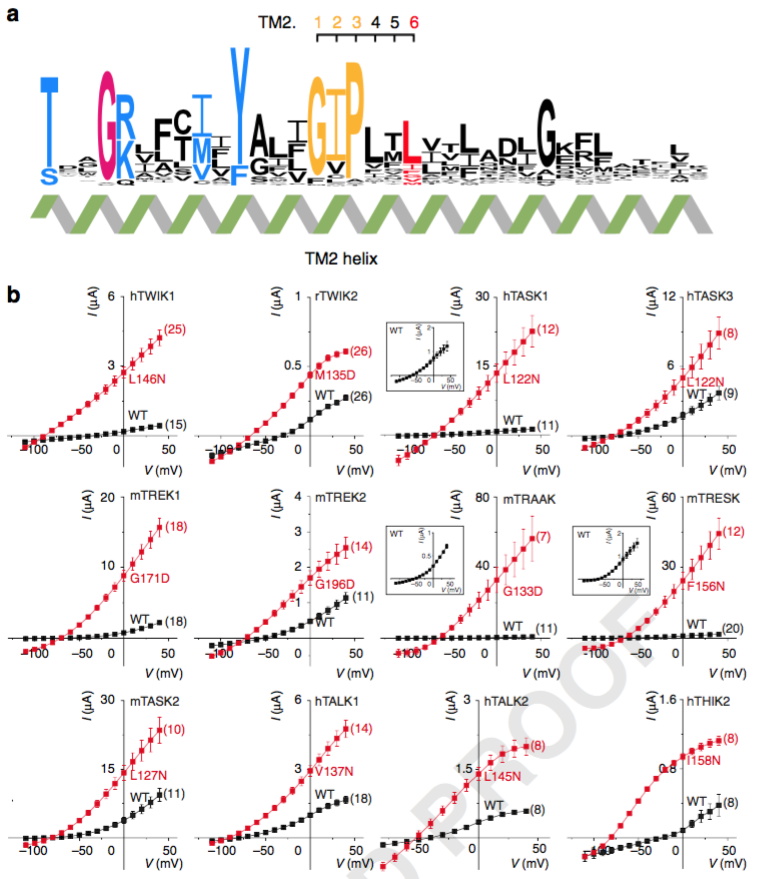
We think this works by increasing K2P channel open probability (first found serendipitously for ORK1 by Ben-Abu, Zhou, Zilberberg and Yifrach in 2009, and by Chatelain et al. and @ProfKalium (S. Tucker) for TWIK1 and THIK channels)
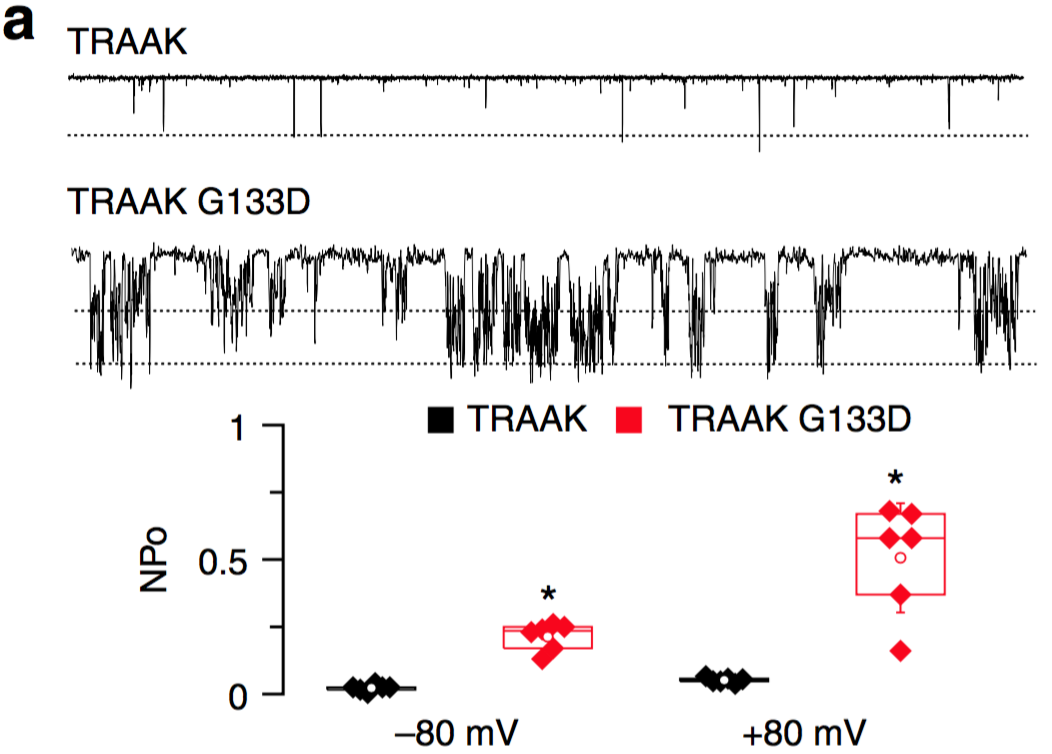
It got better, when we saw that we could tune the effect of these gain-of-function mutants by using different amino acid substitutions.
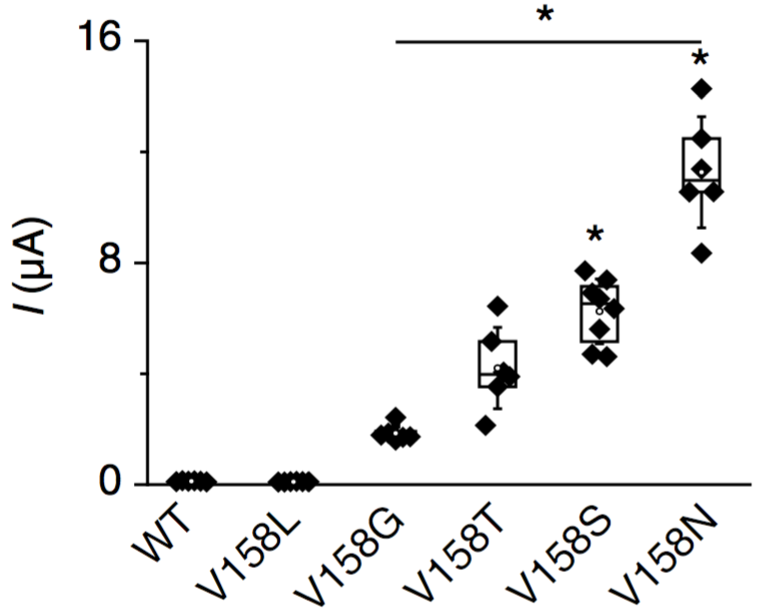
Using CRISPR/Cas9 gene editing, we then created designer gain-of-function mutants in 4 C. elegans K2P channels (and many more since…), which phenocopy those found by Sydney Brenner and Bob Horvitz. This works consistely despite very divergent primary amino acid sequences.
Altoghether, this has been a crucial finding for us because, we were looking for a way to increase the activity of K2P channels in worms to find the genes that are required for their function in vivo.
(On a personnal note, this has solved the entire AIM 3 of our @ERC_Research project *Kelegans*, and then some! Incredibly satisfying when science works out in this way)
And it has opened the way to a (frustratingly) large universe of new projects. Using genetic screens, we have now drawn lines between K2P channels and major cellular players such as Ankyrin, Notch, and most recently, the Dystrophin complex! More about that in the coming year.
Let us know if you have comments, questions, are interested in reagents or collaborations, or just want to chat about our work.
This work was made possible by support from ERC Research (Kelegans), Telethon France (AllianceMyoNeurALP), Agence National de la Recherche, PIA LabexICST, Université Lyon 1, CNRS, and INSERM
Operational!
2018-01-30
All moving boxes are gone. Science going on.



That new lab smell...
2018-01-02
After a rocky move across town featuring: snow, ice, rain, city-wide power outage, failing elevators, missing keys, we are (almost) all setup and operational in our brand spanking new lab space.
14 teams and 200 people finally under/on one roof/floor! Now, let's do some science.


Mind blown...
2017-10-17
What a shock when you turn on the microscope and see such a striking subcellular localisation at the tip of every muscle cell. Figuring out how this is possible is going to be an amazing journey.
Reliable CRISPR/Cas9 genome engineering in Caenorhabditis elegans using a single efficient sgRNA and an easily recognizable phenotype.
Update 2020-12-07
146 Labs and counting! wrmScarlet is freely available for all labs who want it. Visit the following page for the page dedicated to this paper.
To efficiently and freely distribute the wrmScarlet sequence to the community, we have decided to list here all the labs that we have sent the sequence to. Please contact us or anyone on the list to get your copy of the plasmid.
More information here

Great Meeting!
Posters, check! Talk, check! 30 wrmScarlet vectors gone in 3 min, check! 70+ requests to be sent out in the coming days, pending...
And to top it off, Sonia wins the Dr. Matthew J. Buechner Tie Award for service to the community!
July 6, 2017
"Genes to Genomes" announces #Worm17 GSA Poster Award Winners


Finally!
2017-03-02
Our paper has just been accepted at G3: GENES, GENOMES, GENETICS!
27 days from submission to acceptance!
Congratulations to the whole team! It was a great collective effort.